HypoQuantyl
An image processing and analysis tool for automated, high-throughput measurements of hypocotyl growth kinematics.
Refer to the publication resulting from this study:
Separate sites of action for cry1 and phot1 blue-light receptors in the
Arabidopsis hypocotyl
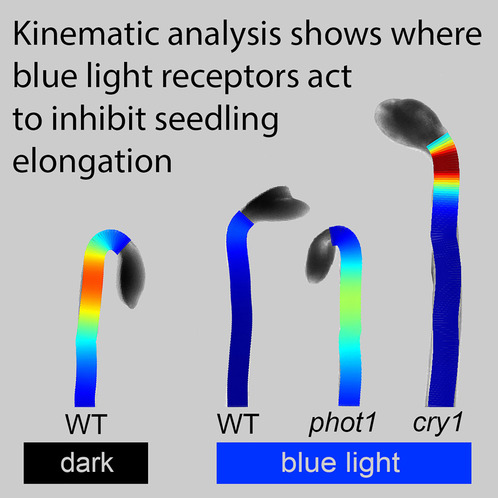
Growth Kinematics
Growth kinematics describes the spatial distribution of growth along a surface. In plants, this refers to the rate of elongation of cells along different regions of the stem. Historically, this has been measured using external markers along the stem’s length. HypoQuantyl achieves this by tracking distinct texture patches along the midline of the hypocotyl over time. These patches, or ‘elements,’ represent stem segments, and their expansion allows the software to calculate growth rates expressed as the Relative Elemental Growth Rate (REGR).
Machine Learning-Based Segmentation
HypoQuantyl provides an automated and robust segmentation process for each frame in a time series of images and is capable of handling multiple well- separated seedlings per image. The machine learning-based segmentation pipeline generates contours of the upper hypocotyl, resolves cotyledon overlap, and identifies and splits the contour at the hypocotyl-cotyledon junction. The system is self-correcting, ensuring reliable results. From the contour, it generates a midline used to compute REGR.
High-Throughput Capabilities
The automated nature of the processing and segmentation pipeline allows deployment on distributed computing servers provided by HTCondor. Using these servers, we have analyzed over 23,000 individual images in a high-throughput manner.
Getting Started
System Recommendations:
-
This software was optimized for Linux operating systems and has been tested on MacOS. We have not yet tested on Windows. There may be some places that do not handle the differences in file path conventions (using ‘/’ in Linux/Mac vs ‘
’ in Windows). -
If running with
parallelflag (par = 1), I recommend a machine with >12 CPU cores to ensure stability. -
8+ GB RAM minimum, and 24+ GB if using
parallelflag. -
MATLAB Version:
This tool was developed on Matlab R2018a to R2022b. Versions slightly earlier and later should work fine but are not guaranteed.
MATLAB Toolboxes:
- Curve Fitting Toolbox
- Global Optimization Toolbox
- Image Processing Toolbox
- Mapping Toolbox
- Sensor Fusion and Tracking Toolbox
- Signal Processing Toolbox
- Statistics and Machine Learning Toolbox
- Symbolic Math Toolbox
- Wavelet Toolbox
(Optional) if you run pipeline with parallel flag (par = 1).
-
Install the required MATLAB toolboxes listed above. Verify your toolboxes using using the following Matlab console command:
matlab.addons.installedAddons;
Download Neural Net Models
- Download the
HQ.matfile from our lab’s Dryad data repository. This contains neural net models, PCA eigenvectors, helper functions, and constants required to run this program [64 MB].
Download Sample Image Datasets
-
Download example image stacks from the same Dryad repository. See below in Pipeline Overview for details about filename conventions.
- The sample data are time-lapse image stacks of the following:
- [
single_seedling.zip] Single cry1 mutant seedling grown for 8 h in darkness [96 MB] - [
multiple_seedling.zip] Five wt seedlings grown for 6 h in blue light, after an initial 2 h in darkness [20 MB]
single_seedling.zip

multiple_seedling.zip

- [
-
Place image stacks and
HQ.matinto the same directory. Make sure this code repository is in a separate directory.mkdir analysis_folder; mv single_seedling.zip analysis_folder; mv multiple_seedling.zip analysis_folder; mv HQ.mat analysis_folder; cd analysis_folder; unzip single_seedling.zip unzip multiple_seedling.zipClone this repository:
git clone https://github.com/jbustamante35/hypoquantyl.git
- Add
hypoquantylto your MATLAB path (with subfolders)- NOTE! remember to remove the
./.gitsubfolder to avoid messy paths
- NOTE! remember to remove the
- The sample data are time-lapse image stacks of the following:
Running HypoQuantyl
-
Open hypoquantyl_script.m in the matlab editor.
edit hypoquantyl_script; -
In the script, set general options as needed:
(1) [tset] Sample image stack to analyze
(2) [vrb] Verbosity
(3) [sav] Save results into .mat files
(4) [par] Run with parallelization [see above on System Requirements],
(5) [odir] File path to output directory
(6) [edate] Date of analysishypoquantyl_script.m%% HypoQuantyl Parmeters Script % 1) Select sample data to run. Uncomment as needed. tset = 'single'; % [single_seedling.zip] 1 cry1 mutant grown for 8h in darkness % tset = 'multiple'; % [multiple_seedling.zip] 5 wt seedlings grown for 2h in darkness then 6h blue light % 2) File path to where you downloaded and unzipped the sample images % path_to_data = '/home/username/Downloads/testimages'; % 3) General options vrb = 1; % Verbosity [ 0 none | 1 verbose ] sav = 1; % Save results into .mat files par = 0; % Use parallel processing [0 | 1] odir = pwd; % Directory path to store results [default pwd] % Advanced parameters are below, but not recommended to toggle unless you know % how they are implemented in this pipeline -
Run
HypoQuantyl.min the Matlab console!- A testing script is provided in tests/hypoquantyl_testing_script.m.
Or simply run:
HQ = HypoQuantyl; -
Analysis of Results [to-do]
- Results are stored in the location set as the output directory (
odir).
- Results are stored in the location set as the output directory (
NOTE ABOUT PERFORMANCE
Runtimes may vary drastically depending on processing power, total CPU threads, and use of the parallel processing parameter.
On our CENT OS RHEL 7 Linux server with an Intel Xeon Silver 4210R CPU of 40 cores, analyzing a single seedling for 96 frames takes about 10 min.
On a conventional personal laptop with 8 cores and no use of parallel processing, a single seedling may take over 5 hours.
Pipeline Overview:
- Input: High-resolution time-lapse image sequence of A. thaliana
hypocotyls.
- Image resolution must clearly capture natural textures along the seedling.
- Growth between frames should not exceed 20 pixels for accurate tracking.*
- Filename convention: condition_genotype_tframe.TIF
- Example:
blue_cry1_t020.TIFrefers to the 20th frame of a cry1 mutant under blue light conditions.
- Example:
- Directory structure:
condition/genotype/imagestacks/imageframes/- Example:
.../multiple/dark/cry1/230220_dark_cry1/dark_cry1_t001.TIF
##### Sample Movie

- Image Processing: Grayscale thresholding and basic object detection to
prepare the seedlings for segmentation.
- Hypocotyls are isolated and split into upper and lower regions. **
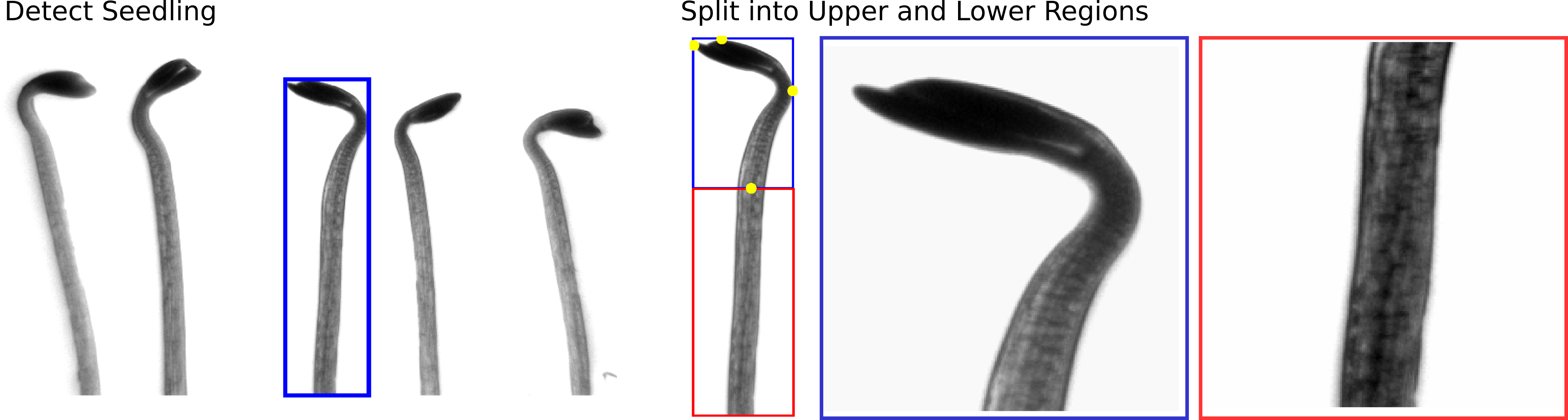
[to-do] finish pipeline descriptions
- Segmentation Pipeline: A 3-stage machine learning pipeline generates the
midline used to measure growth kinematics.
- S-Phase: A ‘seeding’ phase where a convolutional neural network (CNN) initializes the grayscale image with reference frames.
- C-Cycle: A recursive feed-forward neural network loop that predicts ‘Contour’ points based on the S-Phase reference frames and image data.
- R-Cycle: A refinement loop that evaluates the output through a minimization function. The ‘grade’ is the probability that the output is within bounds of the ground truth dataset. If the threshold isn’t met, the process is retried with adjustments.
- REGR Measurements: Calculates the Relative Elemental Growth Rate (REGR) -
- Expressed as a %/hour to describe how ‘elements’ moved away from the top or apex of the seedling.
* 20 pixels is the threshold used for our purposes, but the actual limit may be higher.
** The segmentation pipeline processes only the upper region; lower regions are segmented using basic grayscale thresholding.
Authors
Julian Bustamante, PhD Researcher (jbustamante@wisc.edu)
University of Wisconsin - Madison, Department of Botany
Nathan Miller, Senior Scientist (ndmill@gmail.com)
University of Wisconsin - Madison, Department of Botany
Acknowledgements
Guosheng Wu: Generated the original image dataset used to train early machine learning models.
License
MIT license can be found in the LICENSE file.